2019-08-23
Data Science Command-Line Tools
Description of GNU utils and other less standard tools that helps with processing data from CLI or with shell scripts.
There are plenty of tools designed to ease the life of a Data Scientist. In this group, the special place has tools that are used from the command line. They are special because are available for most operating systems and are designed with Unix philosophy in mind: they do one thing extremely well, they can be chained creating convenient workflows.
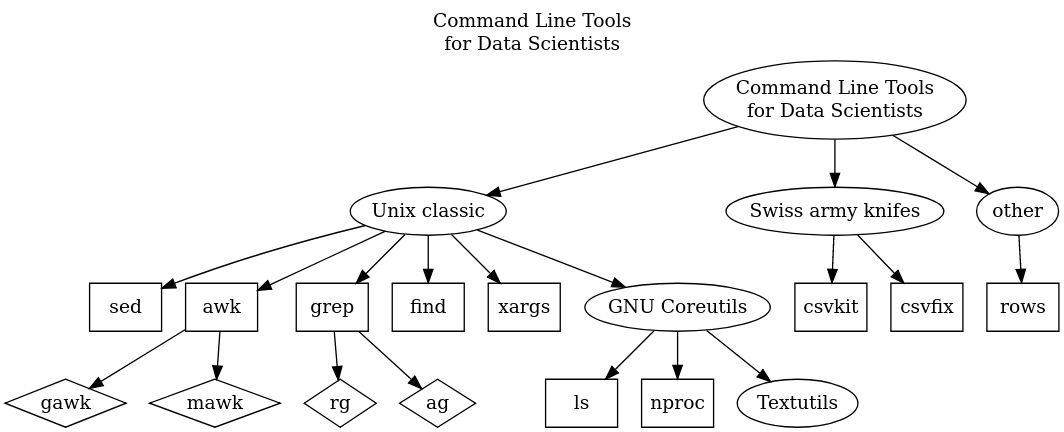
Textutils from GNU Coreutils
From my experience, I have benefited most from mastering the GNU Coreutils. This is the collection of shellutils, fileutils, and textutils - and in this post, I will be discussing the latter.
Table 1. Textutils - text processing tools, part of GNU Coreutils
| command | description |
|---|---|
| cat | merge, print files to standard output |
| comm | compare sorted files line by line; can find differences, unique lines for each compared files |
| csplit | divide files into parts using context |
| cut | remove sections from each line in the file |
| expand/unexpand | convert tab into spaces / convert spaces to tabs |
| fmt | simple text formatter |
| fold | fold each input line to fit the given line length |
| head/tail/shuf | display starting lines of file / display file ending lines/get random lines from file |
| join | join lines from two files on common fields |
| nl | line numbering |
| paste | merge lines |
| sort | sort lines of the text file |
| split | divide file into parts |
| tac | merge, print in reverse order lines of files to standard output |
| tr | translate or remove characters |
| uniq | remove duplicated lines from the file |
| wc | display the number of characters, words, and lines of the given file |
I will give a few examples of commands I use most often or ones that are exceptionally useful.
comm
The comm command is very handy when comes to comparing lists stored in separate files. For example, you have a list of samples used for experiment #1 in one file and a list of samples used in experiment #2 in another file.
The comm command takes sorted files as input and calculates unique lines for FILE1 and for FILE2 and the result is represented respectively as column 1 and column 2. Having unique lines in column 1 and 2, the set of lines that appear in both files is calculated and the result is represented by column 3.
Columns description for comm:
| 1 | 2 | 3 |
|---|---|---|
| lines unique to FILE1 | lines unique to FILE1 | lines that appear in both files |
Having three columns calculated we can now suppress lines that appear in the given column in order to get lines that fulfill the desired condition.
Meaning of the numbers used as comm parameters:
-1suppress column 1 (lines unique to FILE1)-2suppress column 2 (lines unique to FILE2)-3suppress column 3 (lines that appear in both files)
See below three typical usages of comm:
# find lines only in file1
comm -23 file1 file2
# find lines only in file2
comm -13 file1 file2
# find lines common to both files
comm -12 file1 file2
head
By default, head prints the first 10 lines of each file provided as an argument to standard output. With more than one file, precede each with a header giving the file name. You can control how many lines are displayed with option -n:
head -n5 file.txt
You can also print all but the last N lines by adding - sign. For example in order to have all lines but the last one:
head -n -1 file.txt
shuf
To quickly inspect the content of the dataset in a text file you can use head which shows some first lines of the file. If you would like to have a more representative example of the content of the file, you can take a random sample of lines from the file with shuf:
shuf -n 5 file.txt
sort
Here are options I often use for sort and unique operation:
-b, --ignore-leading-blanks
ignore leading blanks
-u, --unique
with -c, check for strict ordering; without -c, output only the first of an equal run
-f, --ignore-case
fold lower case to upper case characters
-i, --ignore-nonprinting
consider only printable characters
sort -bufi data.csv
split
Sometimes there is a need to split datasets into smaller parts. For E.g. when processing large files you can be hit by memory limitations, or you want to speed up processing using parallel computing. To split the file into N parts with the equal number of lines, use:
split -n l/N data.csv
e.g.
split -n l/10 data.csv
This will create series of files named: xaa, xab, xac,.... The default pattern of split for file naming is PREFIXaa, PREFIXab,...; default PREFIX is x. You can provide your own prefix e.g.
tail
split l/10 data.csv part_
and this will result in having files part_aa, part_ab,.... Suffixes don't have to be alphabetical: can be numeric (use switch -d) or even hex (use switch -x). What is particularly useful - prefixes can be used to save resulting files in a given location. Let's take an example dataset.csv file located in the data directory, we want to split this file into parts and save results in parts directory as shown below:
data
├── dataset.csv
└── parts
├── xaa
├── xab
└── xac
To achieve this create a parts directory and modify the prefix to parts/x:
split l/3 data.csv parts/x
Skipping the header row can be done with the tail
tail -n +2 file.txt
This syntax with the usage of + sign might require explanation. Here is an excerpt from the man page:
-n N means output the last N lines, instead of the last 10; or use +N to output lines starting with the Nth.
The usage of + sign can be considered as inverting the argument and telling the tail to print everything but the first x-1 lines. Note that tail -n +1 would print the whole file, tail -n +2 everything but the first line, etc.
the alternative solution involves sed:
sed 1d file.txt
uniq
often used with sort in a way:
cat file.txt | sort | uniq
If there are no special circumstances cat should be avoided:
sort file.txt | uniq
Moreover, sort has the option -u which stands for unique. Therefore, to have unique lines it is sufficient to use:
sort -u file.txt
wc
- count lines
wc -l data.csv
- count lines in multiple files at once
wc -l *.csv
For special tasks - counting lines in very large files one can consider using replacement: Super-Fast Multi-Threaded Line Counter
Any comments or suggestions? Let me know.
To cite this article:
@article{Saf2019Data,
author = {Krystian Safjan},
title = {Data Science Command-Line Tools},
journal = {Krystian's Safjan Blog},
year = {2019},
}
Tags:
machine-learning
Linux